112年度GIS專題中心暑期實習生成果介紹(6)
專題題目:Biomedical Relation Extraction
實習學生:Wilailack Meesawad(國立中央大學人工智慧國際碩士學位學程)
指導老師:蔡宗翰 研究員
Wilailack Meesawad indicates that Biomedical relation extraction plays a critical role in many downstream applications, such as drug discovery and personalized medicine. Most existing datasets focus only on relations between two entities and within single sentences. However, a biomedical relation is commonly expressed in multiple sentences and the same document describes relations between multiple biomedical entities. Due to the reason, she develop NLP system to extract asserted relationships from biomedical text and classifies relations that are novel findings as opposed to background or other existing knowledges that can be found elsewhere.
1.Dataset
Wilailack Meesawad uses the dataset of the BioRED corpus. A Biomedical Relation Extraction Dataset with multiple entity types (e.g., gene/protein, disease, chemical) and relation pairs (e.g., gene-disease; chemical-chemical) at the document level, on a set of 600 PubMed abstracts (accepted by National Center for Biotechnology Information, NCBI).
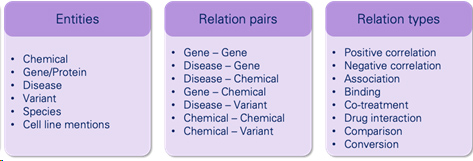
2.Methodology
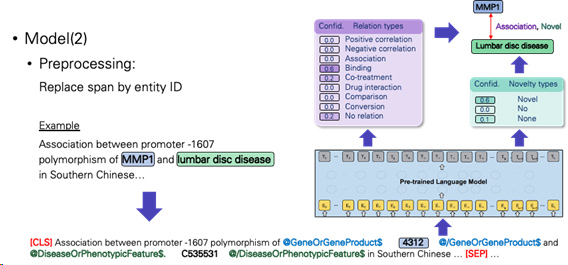
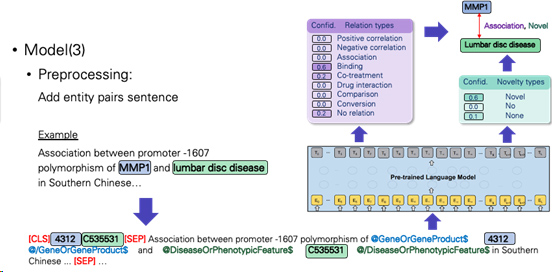
a.Pair the biomedical entity with text and its label. Treat this task as a text classification task with transformer architecture.
-Add entity type before and after span
-Experiment with four pretrained language model:
- PubMedBERT (Gu et al., 2020)
- BioELECTRA (Kanakarajan et al., 2021)
- BioALBERT (Naseem et al., 2020)
- Bioformer (Chen et al., 2021)
-Hyper-parameter setting:

-Results:
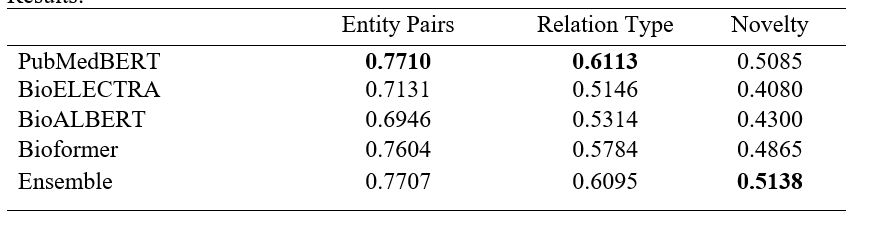
The results show that PubmMedBERT perform best comparing to different pretrained language model (except for novelty score which only slightly lower than ensemble for all pretrained language model). As a result, Wilailack Meesawad choose PubMedBERT pretrained language model to experiment with different model input formats.
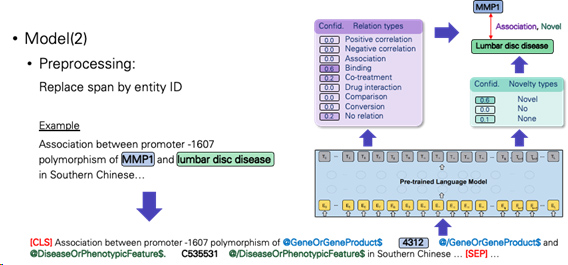
b.Experiment with three different model input formats.
-Replace span by entity ID
-Add entity pairs sentence
-Pretrained language model:
- PubMedBERT (Gu et al., 2020)
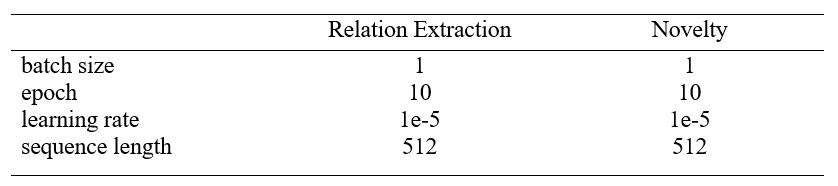
-Hyper-parameter setting:
-Results:
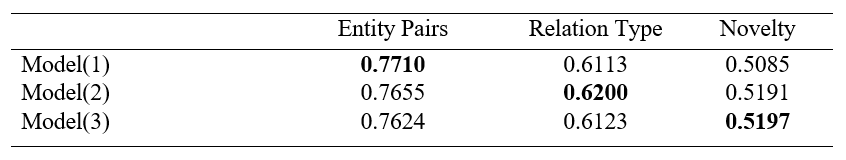
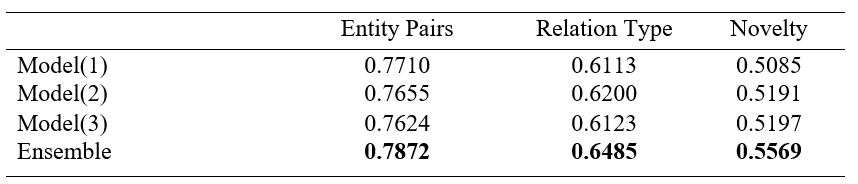
The results show that different model input formats can capture different features and perform well on different benchmark. Model(1) can capture well on entity pairs benchmark while Model(2) and Model(3) can capture well on relation type and novelty benchmark respectively.
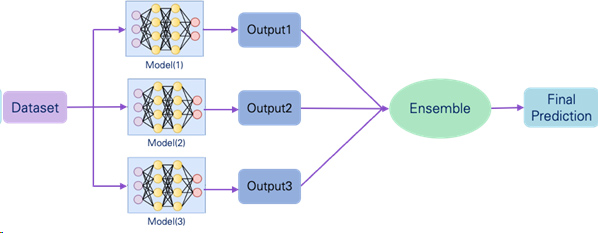
c.Apply ensemble learning method to boost model performance.
-As a result, different model input formats capture different feature and perform well on different benchmark (entity pairs, relation type, and novelty). So, the research apply ensemble learning to boost the model performance.
-the research apply Max Rule ensemble learning method relies on the probability distributions generated by each classifier.
– Results:
3.Limitations and Future work:
Due to the limited resources for example GPU memory, it would take long time to experiment. To prevent parameter inefficient, in future work, Wilailack Meesawad would like to apply transfer learning with adapter modules. By applying adapter, it can reduce the trainning time and resource requirements. Moverover, it allow us to fine-tuning the large language model (e.g., LLaMa (Touvron 2023).
